Authors: Joppe Geluykens, Daniel Treiman, Connor McCormick, Arnav Garg, Travis Addair, Geoffrey Angus, Julian Bright, Jim Thompson.
Editors: Justin Zhao, Daliana Liu, Piero Molino.
Overview
Ludwig is an open-source declarative machine learning framework that enables you to train and deploy state-of-the-art tabular, natural language processing, and computer vision models by just writing a configuration.
Earlier this year, we migrated our entire backend from TensorFlow to PyTorch. In this latest release, we’ve capitalized on the benefits of being on a fully PyTorch backend — adding Gradient Boosted Models (GBMs), expanding options for deployment with Pipelined TorchScript and NVIDIA Triton, all while continuing to improve testing and stability with parameter update tests and config validation.
All of this, in addition to support for Ray 2.0, makes Ludwig 0.6 a great platform for machine learning, supporting a variety of state-of-the-art ML models combined with MLOps best practices packaged in the form of a simple configuration interface.
Ludwig 0.6 feature highlights:
- Gradient boosted models: Historically, Ludwig has been built around a single, flexible neural network architecture called
ECD(for Encoder-Combiner-Decoder). With the release of 0.6 we are adding support an additional model architecture: gradient-boosted tree models (GBMs). - Configuration type checking and schema validation: We’ve defined a formalized Ludwig configuration schema that validates configurations at initialization time, which can help you avoid mistakes like typos and syntax errors.
- Calibrating probabilities for category and binary output features: With deep neural networks, probabilities given by models often don’t match the true likelihood of the data. Ludwig now supports temperature scaling calibration (On Calibration of Modern Neural Networks), which brings class probabilities closer to their true likelihoods in the validation set.
- Pipelined TorchScript: We improved the TorchScript model export functionality, making it easier than ever to train and deploy models for high performance inference.
- Model parameter update unit tests: The code to update parameters of deep neural networks is complex for developers to make sure the model parameters are updated. To address this difficulty and improve the robustness of our models, we implemented a reusable utility to ensure parameters are updated during one cycle of a forward-pass / backward-pass / optimizer step.
Additional improvements include a new global configuration section, time-based dataset splitting and more flexible hyperparameter optimization configurations. Read more about specific features below!
If you are learning about Ludwig for the first time, or if these new features are relevant and exciting to your research or application, we’d love to hear from you. Join our Ludwig Community.
Gradient Boosted Models
Historically, Ludwig has been built around a single, flexible neural network architecture called ECD (for Encoder-Combiner-Decoder). With the release of 0.6 we are, adding support an additional model architecture: gradient-boosted tree models (GBM).
This is motivated by the fact that tree models still outperform neural networks on some tabular datasets, and the fact that tree models are generally less compute-intensive, making them a better choice for some applications. In Ludwig, you can now experiment with both neural and tree-based architectures within the same framework, taking advantage of all of the additional functionalities and conveniences that Ludwig offers like: preprocessing, hyperparameter optimization, integration with different backends (local, ray, horovod), and interoperability with different data sources (pandas, dask, modin).
How to use GBMs?
Install the tree extra package with pip install ludwig[tree]. After the installation, you can use the new gbm model type in the configuration. Ludwig will default to using the ECD architecture, which can be overridden as follows to use GBM:
model_type: gbm
In some initial benchmarking we found that GBMs are particularly performant on smaller tabular datasets and can sometimes deal better with class imbalance compared to neural networks. Like the ECD neural networks, GBMs can be sensitive to hyperparameter values, and hyperparameter tuning is important to get a well-performing model.
Under the hood, Ludwig uses LightGBM for training gradient-boosted tree models, and the LightGBM trainer parameters can be configured in the trainer section of the configuration. For serving, the LightGBM model is converted to a PyTorch graph using Hummingbird for efficient evaluation and inference.
Limitations
Ludwig’s initial support for GBM is limited to tabular data (binary, categorical and numeric features) with a single output feature target.
Calibrating probabilities for category and binary output features
Suppose your model outputs a class probability of 90%. Is there a 90% chance that the model prediction is correct? Do the probabilities given by your model match the true likelihood of the data? With deep neural networks, they often don’t.
Drawing on the methods described in On Calibration of Modern Neural Networks (Chuan Guo, Geoff Pleiss, Yu Sun, Kilian Q. Weinberger), Ludwig now supports temperature scaling for binary and category output features. Temperature scaling brings a model’s output probabilities closer to the true likelihood while preserving the same accuracy and top k predictions.
How to use Calibration
To enable calibration, add calibration: true to any binary or category output feature configuration:
output_features:
- name: Cover_Type
type: category
calibration: trueWith calibration enabled, Ludwig will find a scale factor (temperature) which will bring the class probabilities closer to their true likelihoods in the validation set. The calibration scale factor is determined in a short phase after training is complete. If no validation split is provided, the training set is used instead.
To visualize the effects of calibration in Ludwig, you can use Calibration Plots, which bin the data based on model probability and plot the model probability (X) versus observed (Y) for each bin (see code examples).
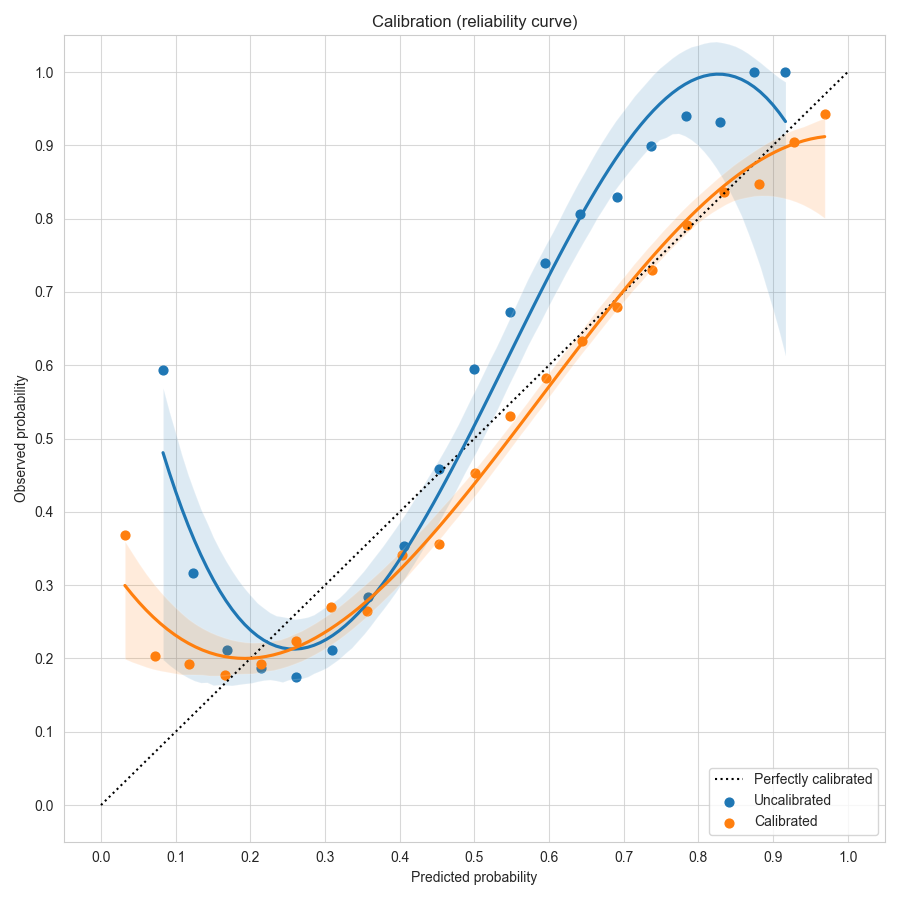
In a perfectly calibrated model, the observed probability equals the predicted probability, and all predictions will land on the dotted line y=x. In this example using the forest cover dataset, the uncalibrated model in blue gives over-confident predictions near the left and right edges close to probability values of 0 or 1. Temperature scaling learns a scale factor of 0.51 which improves the calibration curve in orange, moving it closer to y=x.
Limitations
Calibration is currently limited to models with binary and category output features.
Config type checking and schema validation
Ludwig configurations are flexible by design, as they internally map to Python function signatures. This allows for expressive configurations, but we have found that users would too easily have typos in their configs like incorrect value types or other syntactical inconsistencies.
We have now formalized the Ludwig config with a strongly typed schema, which also serves as a centralized source of truth for parameter documentation and config validation. Ludwig configurations are now rigorously checked for valid types and values, flagging issues early at model initialization time.
Nested encoder and decoder parameters
We have also restructured the way that encoders and decoders are configured to now use a nested structure, consistent with other modules in Ludwig such as combiners and loss.
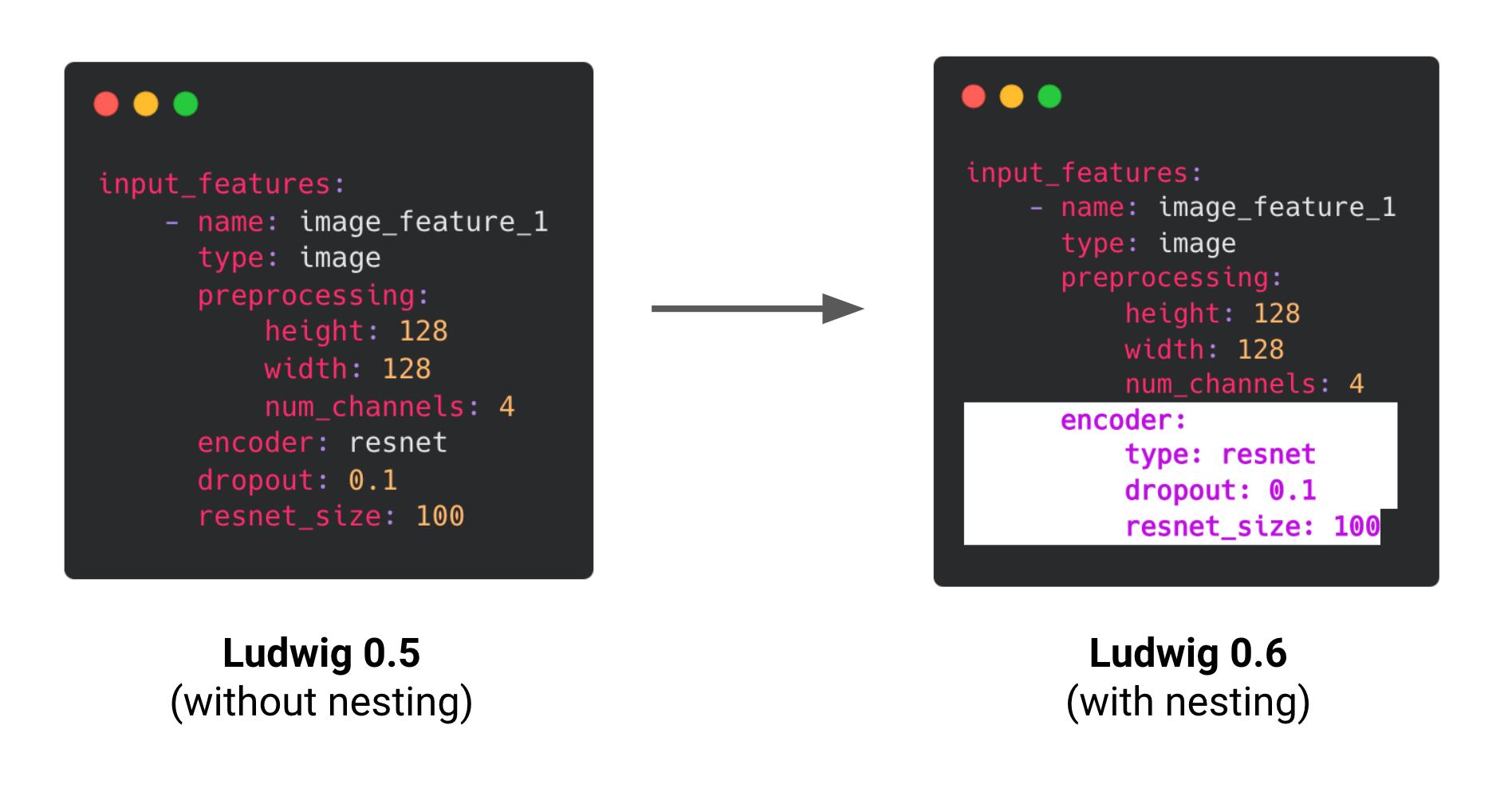
As these changes impact what constitutes a valid Ludwig config, we introduced a registration mechanism to ensure backward compatibility with older versions of Ludwig. Older configs are invisibly and automatically upgraded to the latest config structure.
We hope that with the new Ludwig schema and the improved encoder/decoder nesting structure, you find using Ludwig to be a much more robust and user-friendly experience! New Defaults Ludwig Section
In older versions of Ludwig, you can specify preprocessing parameters on a per-feature-type basis through the global preprocessing section in Ludwig configs. This is useful for applying certain transformations to their data for every feature of the same type. However, there was no equivalent mechanism for global encoder, decoder or loss related parameters.
In Ludwig 0.6, we are introducing a new defaults section within the Ludwig config to define all feature-type defaults for preprocessing, encoders, decoders, and loss. Default preprocessing and encoder configurations will be applied to all input_features of that feature type, while decoder and loss configurations will be applied to all output_features of that feature type.
Note that you can still specify feature specific parameters as usual, and these will override any default parameter values that come from the global defaults section.
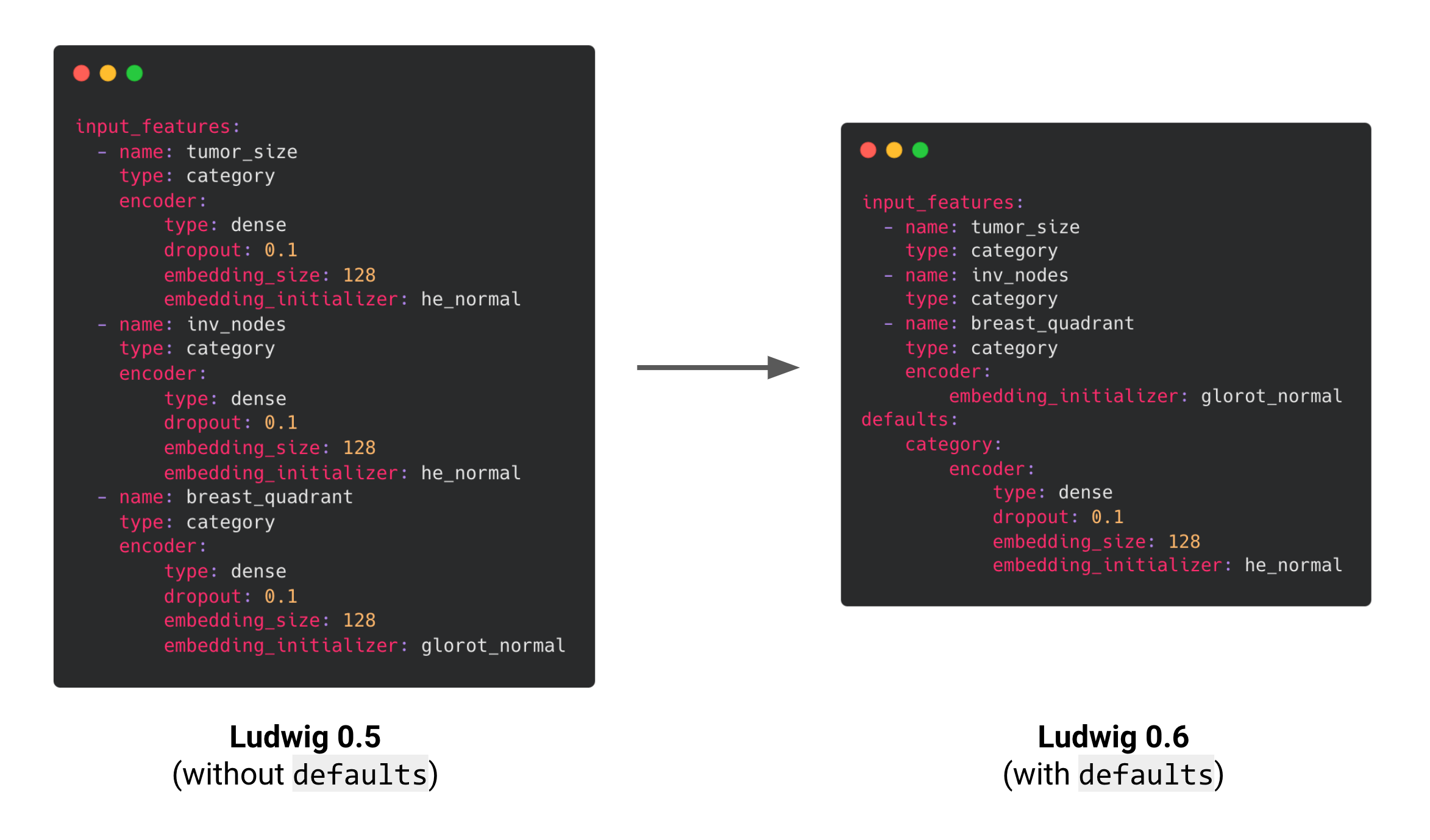
Example of a config of a mammography dataset to predict breast cancer that contains many categorical features. In Ludwig 0.6, common encoder parameters can be declared once in the config’s ‘defaults’ section. Here, the encoder defaults for type, dropout and embedding_size are applied to all three categorical features. The he_normal embedding initializer is only applied to tumor_size and inv_nodes since we didn’t specify this parameter in their feature definitions, but breast_quadrant will use the glorot_normal initializer since it will override the value from the defaults section.
Example of a config of a mammography dataset to predict breast cancer that contains many categorical features. In Ludwig 0.6, common encoder parameters can be declared once in the config’s ‘defaults’ section. Here, the encoder defaults for type, dropout and embedding_size are applied to all three categorical features. The he_normal embedding initializer is only applied to tumor_size and inv_nodes since we didn’t specify this parameter in their feature definitions, but breast_quadrant will use the glorot_normal initializer since it will override the value from the defaults section.
The defaults section enables the same fine-grained control as before with the benefit of making your config easier to define and read.
Global Defaults In Hyperopt
The defaults section has also been added to hyperopt, so that you can define feature-type level parameters for individual trials. This makes the definition of the hyperopt search space more convenient, without the need to define individual parameters for each of the features in instances where the dataset has a large number of input or output features.
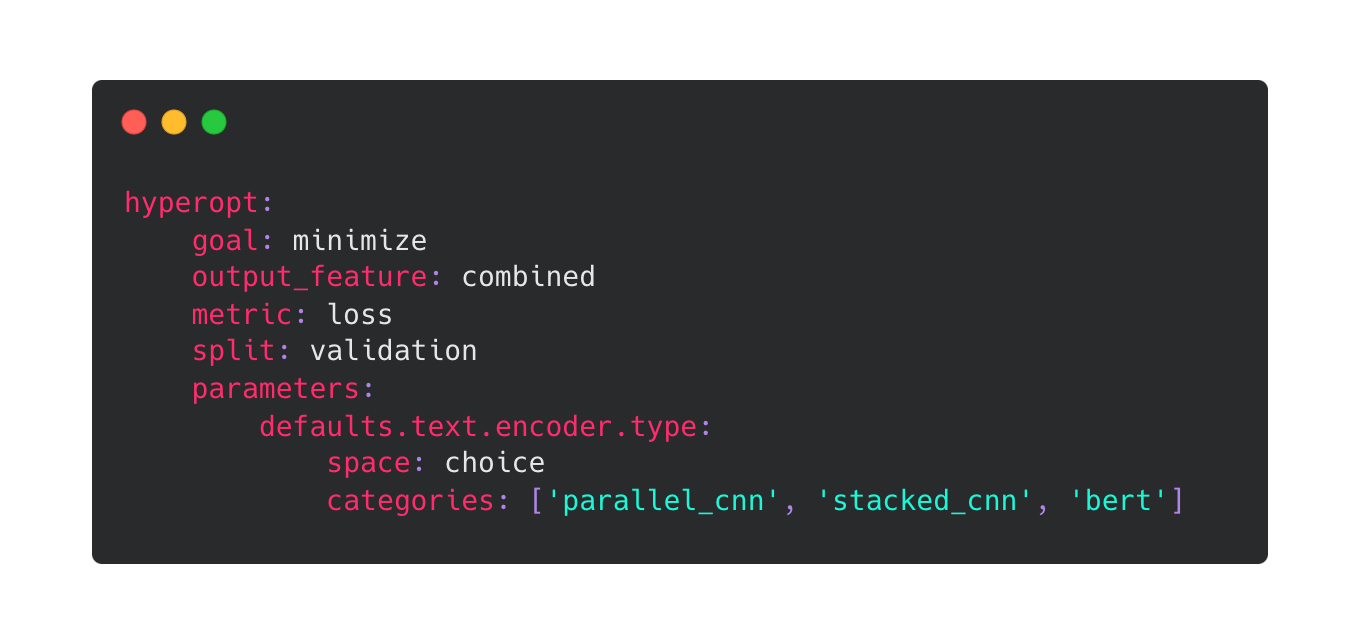
Example of how to specify feature-type global defaults in the hyperopt sub-config. For example, if you want to hyperopt over different encoders for all text features for each of the trials, you can do so by defining a parameter this way. This will sample one of the three encoders for text features and apply it to all the text features for that particular trial.
Configs Blocks In Hyperopt
We have extended the range of hyperopt parameters to support parameter choices that consist of partial or complete blocks of nested Ludwig config sections. This allows you to search over a set of Ludwig configs, as opposed to needing to specify config params individually and search over all combinations.
To provide a parameter that represents a full top-level Ludwig config, the . key name can be used.
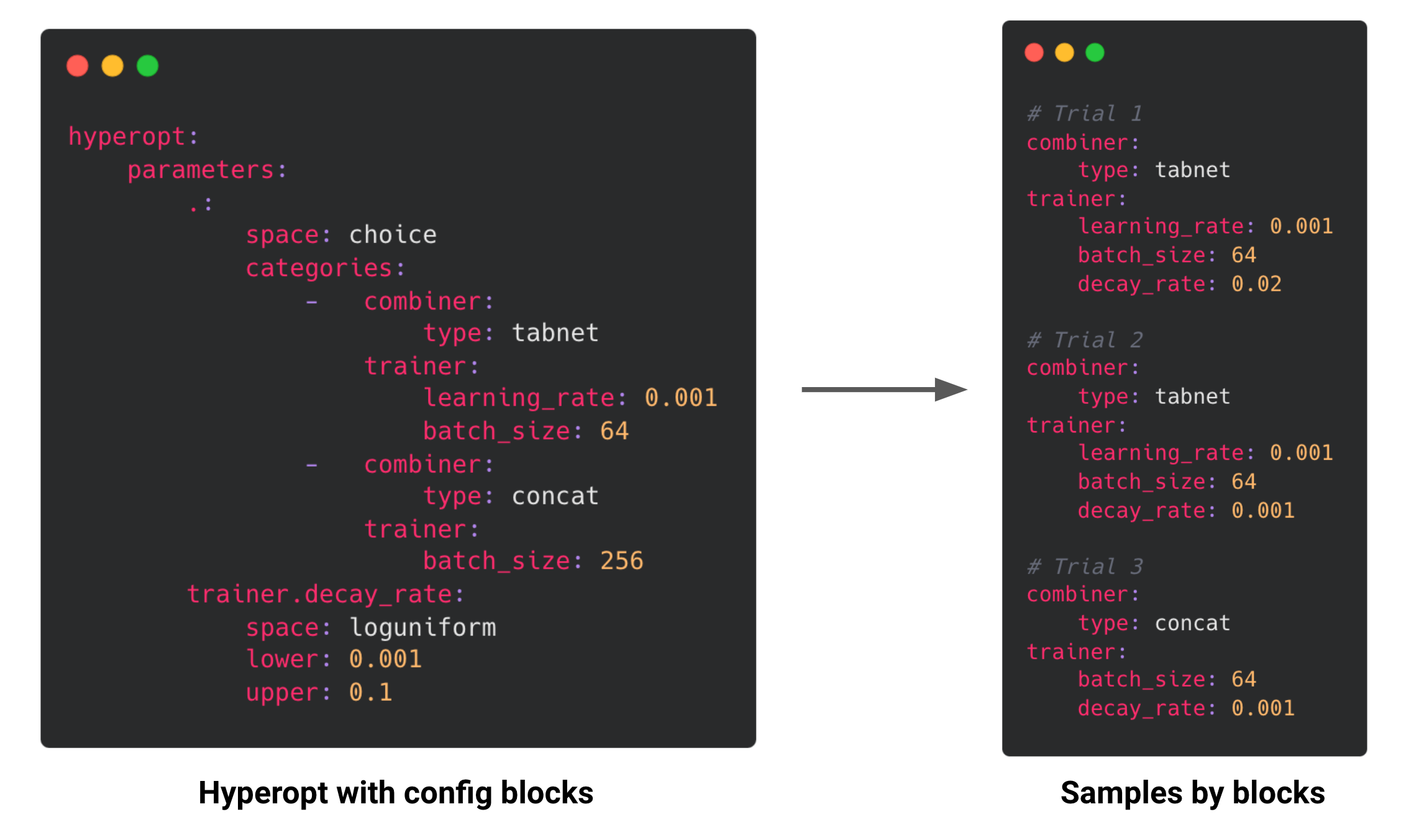
Hyperopt search spaces with configuration blocks are sampled block-wise.
Pipelined TorchScript
In Ludwig 0.6, we improved the TorchScript model export functionality, making it easier than ever to train and deploy models for high performance inference.
At the core of our implementation is a pipeline-based approach to exporting models. After training a Ludwig model, you can run the export_torchscript command in the CLI, or call LudwigModel.save_torchscript. If model training was performed on a GPU device, doing so produces three new TorchScript artifacts:
torchscript/
inference_preprocessor.pt
inference_predictor_cuda.pt
inference_postprocessor.ptThese artifacts represent a single LudwigModel as three modules, each separated by stage: preprocessing, prediction, and postprocessing. These artifacts can be pipelined together using the InferenceModule class method InferenceModule.from_directory, or with some tools such as NVIDIA Triton.
One of the most significant benefits is that TorchScripted models are backend and environment independent and different parts can run on different hardware to maximize throughput. They can be loaded up in either a C++ or Python backend, and in either, minimal dependencies are required to run model inference. Such characteristics ensure that the model itself is both highly portable and backward compatible.
Time-based Dataset Splitting
In Ludwig 0.6, we have added the ability to split based on a date column such that the data is ordered by date (ascending) and then split into train-validation-test along the time dimension. To make this possible, we have reworked the way splitting is handled in the Ludwig configuration to support a dedicated split section:
preprocessing:
split:
type: datetime
column: created_ts
probabilities: [0.7, 0.1, 0.2]In this example, by setting probabilities: [0.7, 0.1, 0.2], the earliest 70% of the data will be used for training, the middle 10% used for validation, and the last 20% used for testing.
This feature is important to support backtesting strategies where you may need to know if a model trained on historical data would have performed well on unseen future data. If we were to use a uniformly random split strategy in these cases, then the model performance may not reflect the model’s ability to generalize well if the data distribution is subject to change over time. For example, imagine a model that is predicting housing prices. If we both train and test on data from around the same time, we may fool ourselves into believing our model has learned something fundamental about housing valuations when in reality it might just be basing its predictions on recent trends in the market (trends that will likely change once the model is put into production). Splitting the training from the test data along the time dimension is one way to avoid this false sense of confidence, by showing how well the model should do on unseen data from the future.
Prior to Ludwig 0.6, the preprocessing configuration supported splitting based on a split column, split probabilities (train-val-test), or stratified splitting based on a category, all of which were flattened into the top-level of the preprocessing section:
preprocessing:
force_split: false
split_probabilities: [0.7, 0.1, 0.2]
stratify: nullThis approach was limiting in that every new split type required reconciling all of the above params and determining how they should interact with the new type. To resolve this complexity, all of the existing split types have been similarly reworked to follow the new structure supported for datetime splitting.
Examples:
Splitting by row at random (default).
preprocessing:
split:
type: random
probabilities: [0.7, 0.1, 0.2]Splitting based on a fixed split column.
preprocessing:
split:
type: fixed
column: splitStratified splits using a chosen stratification category column.
preprocessing:
split:
type: stratify
column: color
probabilities: [0.7, 0.1, 0.2]Be on the lookout as we continue to add additional split strategies in the future to support advanced usage such as bucketed backtesting. If you are interested in these kinds of scenarios, please reach out!
Parameter Update Unit Tests
A significant step was taken in this release to improve the code quality of Ludwig components, e.g., encoders, combiners, and decoders. Deep neural networks have many layers composed of a large number of parameters that must be updated to converge to a solution. Depending on the particular algorithm, the code for updating parameters during training can be quite complex. As a result, it is near impossible for a developer to reason through an analysis that confirms model parameters are updated.
To address this difficulty, we implemented a reusable utility to perform a quick sanity check to ensure parameters, such as tensor weights and biases, are updated during one cycle of a forward-pass / backward-pass / optimizer step. This work was inspired by these earlier blog postings: How to unit test machine learning code and Testing Your PyTorch Models with Torcheck.
This utility was added to unit tests for existing Ludwig components. With this addition, unit tests for Ludwig now ensure the following:
- No run-time exceptions are raised
- Generated output has the correct data type and shape
- (New capability)Model parameters are updated as expected
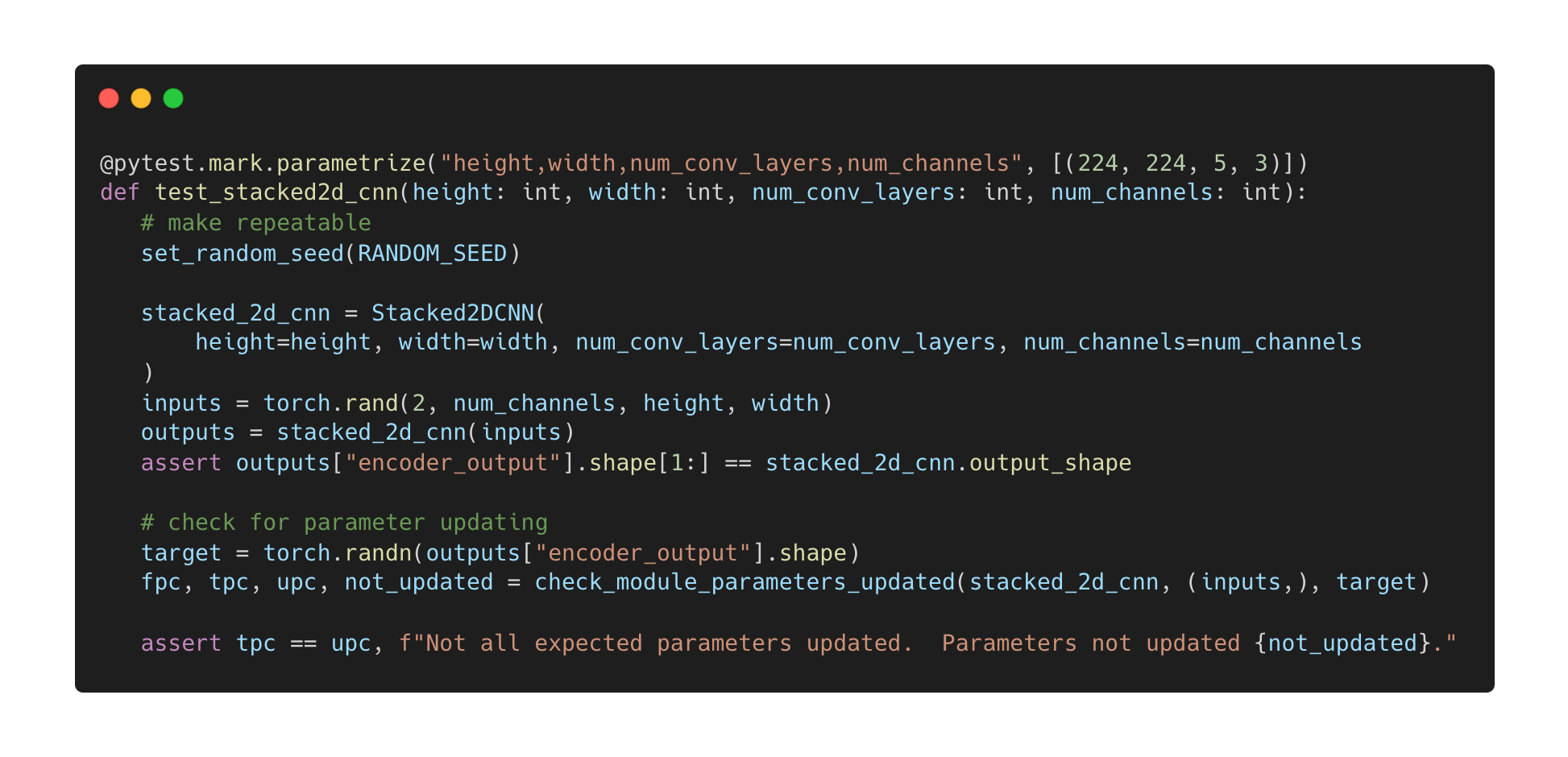
The above is an example of a unit test. First, it sets the random number seed to ensure repeatability. Next, the test instantiates the Ludwig component and processes synthetic data to ensure the component does not raise an error and that the output has the expected shape. Finally, the unit test checks if the parameters are updated under the different combinations of configuration settings.In addition to the new parameter update check utility, Ludwig’s Developer Guide contains instructions for using the utility. This allows contributors developing custom encoders, combiners, or decoders, to ensure the quality of their custom component.
Stay in the loop
Ludwig thriving open source community gathers on Slack, join it to get involved!If you are interested in adopting Ludwig in the enterprise, check out Predibase, the declarative ML platform that connects with your data, manages the training, iteration, and deployment of your models, and makes them available for querying, reducing time to value of machine learning projects.Congratulations to our new contributors!
- @Marvjowa made their first contribution in #2236
- @Dennis-Rall made their first contribution in #2192
- @abidwael made their first contribution in #2263
- @noahlh made their first contribution in #2284
- @jeffkinnison made their first contribution in #2316
- @andife made their first contribution in #2358
- @alberttorosyan made their first contribution in #2413